Christian C. Gruber*, Georg Steinkellner Innophore,...
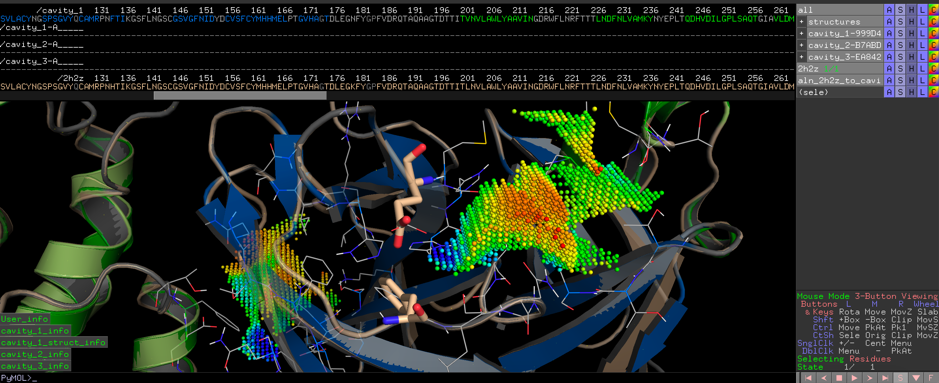
Read MoreResponding to the ongoing epidemic COVID-19 coronavirus outbreak, the FASTCURE consortium aims to rapidly identify, collect and verify chemical compounds and methods suitable for inhibiting SARS-CoV-2 (formerly known as 2019-nCoV) Mpro protease as a primary target.
Worldwide leading institutions active in bioinformatics, drug-screening, and design supported by major European HPC facilities will be redundantly cross-linked with high-throughput experimental in-vitro and in-vivo validation methods comprising cell culture compound screening, X-ray, and NMR complex determination to continuously evaluate potential compounds.
Get more information at www.fastcure.net
Contact us anytime at contributions@fastcure.net